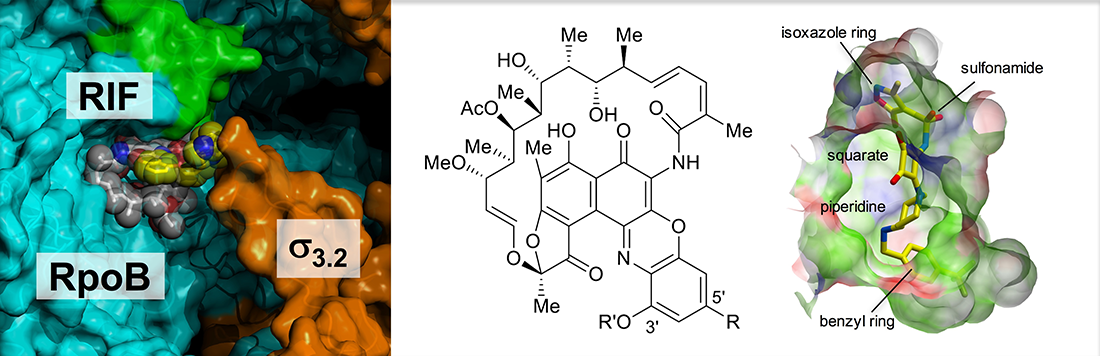
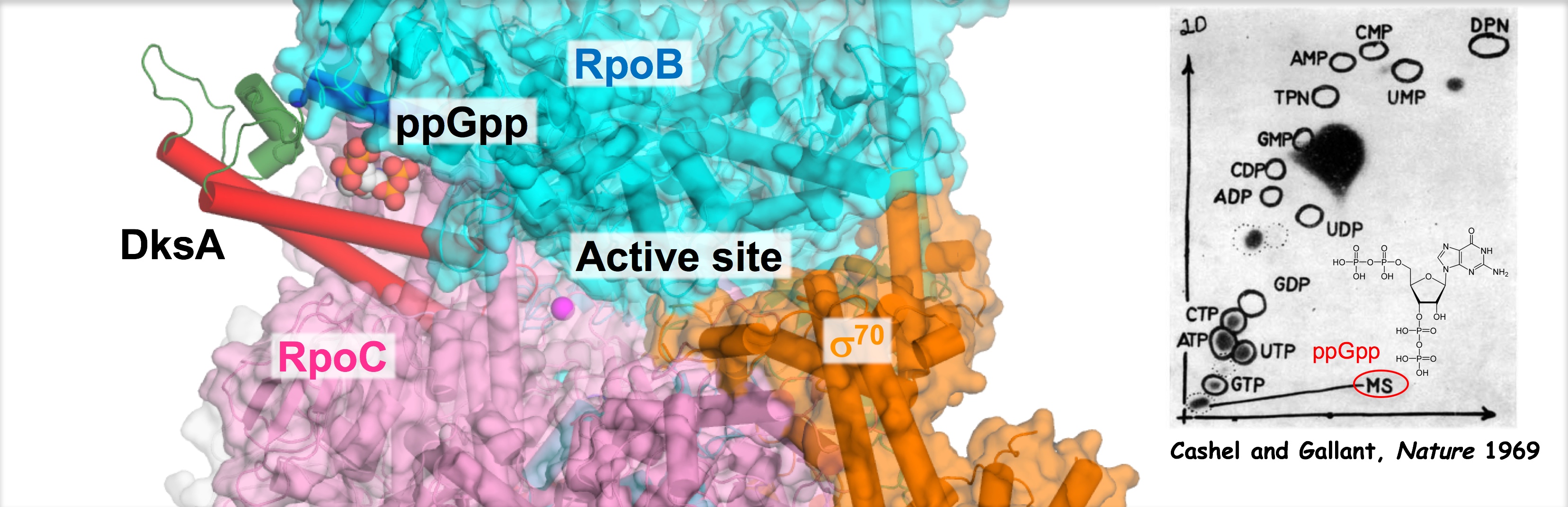
Welcome to the Murakami Lab at Penn State University
Our research focus has been on revealing the molecular mechanisms underlying gene expression and transcription regulation in bacteria, archaea, and eukaryotes as well as SARS_CoV_2.
Open Positions (Postdoc, Graduate and Undergraduate students)
Latest news:
June 11-16, 2023: Manju Narwal, Rishi Vishwakarma and George Fordjour presented their researches at the Gordon Research Conference “Mechanism of Microbial Transcription”
April 10, 2023: A paper reporting the SARS_CoV_2 main protease (Mpro) and polyprotein interaction is published (JBC). Press release (Penn State News).
February 7, 2023: A paper reporting the allosteric mechanism of NusG-dependent transcription pausing is published (PNAS).
January 1, 2023: Natalie Smith (MCIBS) and George Fordjour (BMMB) joined the Murakami lab as graduate students
February 4, 2021: A paper reporting the nucleotide selection mechanism by bacterial RNA polymerase is published (Nature Communications).
January 22, 2021: A paper reporting the cryo-EM study of E. coli RNA polymerase and ribosomal RNA promoter complex is published (Nature Communications). This study showed several new structures for understanding the rRNA transcription expression and regulation, which is vital for life. Press release. Interview.
December 18, 2020: A manuscript reporting the structure and function of HelD, RNA polymerase binding ATP-dependent helicase is published. URee Chon, a former undergrad in my lab determined the X-ray crystal structure of a part of HelD.
November 30, 2020: A manuscript reporting the structure and function of archaeal general transcription factor TFEα is published.
November 25, 2020: Katsu Murakami named a fellow of the American Association for the Advancement of Science (AAAS).